The Integrated Disease Surveillance Programme (IDSP) was established by the National Centre for Disease Control in 2004, as a decentralised, laboratory-based, and IT-enabled disease surveillance system for epidemic-prone diseases. One of the stated objectives of the IDSP, under the aegis of the Ministry of Health and Family Welfare, is to “develop and maintain an Information Communication Technology—for collection, collation, compilation, analysis, and dissemination of real-time data”.
At Wadhwani AI, we use AI to automate the process of disease surveillance. Before we dive deeper into our approach, it is important to first understand the two approaches to disease surveillance currently being followed in India.
One approach has been an indicator-based surveillance system. This involves:
- Healthcare providers reporting to public health officials about specific diseases from units set up in every state.
- Manually collecting data from medical colleges, health centres, hospitals, labs, etc.
- Collecting various types of data, including meteorological data, historical data, and remote sensing inputs.
- Collecting data daily.
- Alerting Rapid Response Teams (RRT) in the event of a rising trend in illnesses in an area.
The second approach, and the one we focus on, is an event-based surveillance system. In India, this has traditionally involved:
- Manual screening of reports, stories, rumours, and other health-related information by the Media Scanning and Verification Cell (MSVC) in New Delhi.
- Analysis of screened information by epidemiologists.
- Disseminating alerts electronically to relevant District Surveillance Officers.
- Distributing verified media alert reports to responsible health authorities and stakeholders for appropriate public health action.
Event-based surveillance systems are very important for the early detection of infectious disease outbreaks. Since the volume of news articles published online is extremely large, manually screening for potential events of interest is not practical. This is the reason most developed countries have automated solutions (ProMED and MedISys, for example), which crawl the web and generate alerts every day1.
In India, the MSVC is responsible for event-based surveillance. Before we visited the MSVC office in February 2022, we had been working on Wadhwani AI’s AI-powered event-based surveillance system for close to four months. We had also created a prototype that we wished to demonstrate to the MSVC for their feedback.
At present, the people working at the MSVC face an overwhelming workload. Their task is to read emails containing the links to articles sent to them via Google Alerts. They also have to read about 20 newspapers daily—10 in English and 10 in Hindi. Afterwards, they generate alerts for the information they find relevant. They vet these alerts with an epidemiologist, and send them to the concerned state authorities by email or through SMS. They also receive follow-ups on alerts sent on previous days from the concerned state authorities, and maintain a record of previously generated alerts.
Carrying out disease surveillance manually is not a trivial task; thousands of news articles are published online daily. Only a handful of these articles contain any relevant information about a potential outbreak. A single person’s oversight could result in a potential outbreak remaining undetected.
This visit reinforced our belief that an automated solution is the only way to carry out event-based surveillance at scale if we want to reap its benefits.
Understanding the problem
A schematic of the overall pipeline of the solution we proposed can be seen in the figure below.
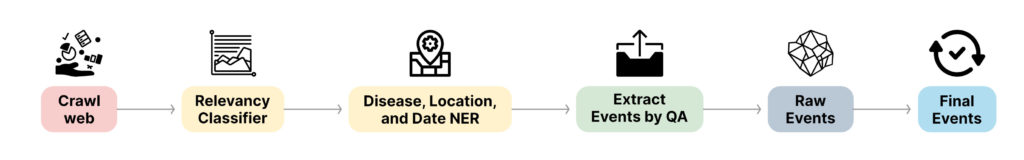
Let’s take a moment to understand the problem at hand. We would like to extract events of interest, related to public health, by analysing news articles, and generate alerts as soon as possible. An event by definition includes the following entities:
- Location
- Disease
- Incident (case or death)
- Incident Type (new or total)
- Number
- Date
Web pages with news articles are typically very long, so to reduce our search space, we rely on the title and description of a news article, as most of the relevant information is summarised in these. Here is an example of what that might look like:
Article: 32 new cases of coronavirus were detected in Delhi, total now at 4028.
Here, there are two events.
Event 1:
Location: Delhi; Disease: Coronavirus; Incident: Case; Incident Type: New; Number: 32
Event 2:
Location: Delhi; Disease: Coronavirus; Incident: Case; Incident Type: Total; Number: 4028
It is common for disease surveillance systems currently deployed around the globe to fetch articles of importance by spotting disease keywords2. If an article mentions the name of a disease—“dengue”, for instance—it will be flagged as important. Given that a lot of news articles containing disease keywords may not actually talk about a disease outbreak, this approach is not the most efficient one. To overcome this problem, some systems also use humans in the loop, whose job is to remove these redundant articles and keep the ones that actually pertain to a disease outbreak.
Our system provides information about the disease outbreak, as well as where it occurred and the number of people affected. This is a significantly more complicated task, leading to richer information and reduced dependency on humans in the loop. In addition to this, while other systems commonly use English as the surveillance language, we have developed this system to work for 10 other Indian languages (Hindi, Marathi, Telugu, Tamil, Malayalam, Punjabi, Gujarati, Oriya, Bengali, Kannada), and our approach can be easily extended to include other languages.
Now that we have understood the problem, let’s look closely at our approach towards a solution.
Crawling the web
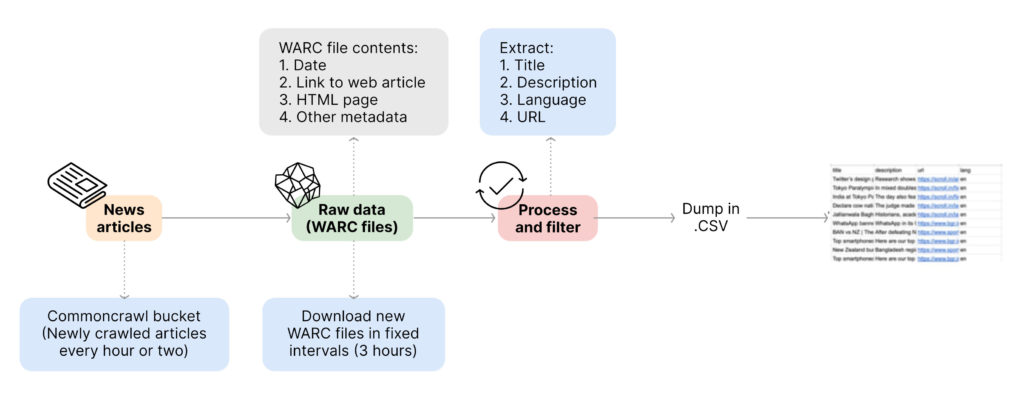
To crawl the web, we take the help of Common Crawl as well as Google Alerts. Common Crawl is a nonprofit organisation that periodically crawls the web for news articles from around the world. It crawls millions of news articles and dumps them in an AWS S3 bucket every hour. We read this dump after every hour or two and process all the new files it adds.
- Since we are only interested in articles related to India, we begin by creating a knowledge base of all the news domains that regularly write about disease outbreaks in India.
- Then, we filter the news articles based on these domain names to narrow down our search space.
- We extract the title, description, language, and URL of these news articles.
- We also use language identification models and filter out languages not supported by our system.
Google Alerts is a service from Google that alerts users as soon as specific keywords we are interested in appear on the web. It serves as the second source of information for our system. We have created a knowledge-base of all the diseases that we want to track, along with any relevant synonyms, and then set up Google Alerts for all these keywords. We receive the news articles which contain these keywords via email, which we then crawl to extract the relevant information, while applying the same domain and language filters as mentioned above.
Both Common Crawl and Google Alerts provide us with millions of fresh news articles daily. Of these articles, we narrow our search down to about 35–40,000 articles by applying domain and language filters.
Relevance classification
General health news makes up a very small portion of the news published online. Most news articles published contain information about sports, entertainment, and politics. To carry out event extraction efficiently, it is important that we filter these articles and keep only the ones that are actually relevant to public health.
Data
Datasets that enable this task are not readily available. There are a few datasets which contain articles related only to COVID-19, such as the data from inshorts and Aylien’s COVID-19 dataset, but our goal was to build a system that works for 40+ diseases, including dengue, malaria, and chickenpox. To solve this problem, we used data augmentation to create data for other diseases.
We first created a knowledge-base of diseases that contains disease names and their synonyms, and used this to replace “COVID-19” and its synonyms with other diseases. These became our positive data (articles related to public health). For negative samples, we used the HuffPost news dataset.
The dataset we prepared consisted of 160,000 news articles in all, out of which around 80,000 were positive articles and the remaining were negative articles. And since this data was in English, we translated it to other languages to train models in those languages.
Model
For articles in English, we used BioBert3, which is a model pre-trained on biomedical texts, and fine-tuned this using the dataset we prepared. Our aim here was to classify whether the given article is relevant to public health. For other languages, we made use of multilingual models like XLM-R4.
This was done to narrow down our search space. Even after being classified as relevant, articles go through more filters further down the pipeline. This is why tagging irrelevant articles as relevant is not as harmful as the removal of actually relevant articles. Our focus while training, therefore, is to improve recall.
A byproduct of this strategy is reduced precision. In practice, by using this model, we are able to reduce the total number of articles by a factor of 3 to 4. We are still left with several irrelevant articles, but we handle them with more filters down the line.
Disease named entity recognition (Disease NER)
Knowing which disease is present in a given article helps us in two ways. First, as mentioned earlier, while we filter articles based on their relevance to public health, we still want to remove all the false positives we are left with. We filter the articles which do not mention any disease. Second, we use the disease information as an anchor to perform event extraction (more on this below).
There are two major requirements our disease NER system must be able to fulfil:
- It should be able to spot known diseases almost perfectly.
- It should be able to spot unknown diseases based on the available context, thus making it future-proof.
For this, we make use of both classical and deep learning methods of doing NER.
Classical approach: To perform string-matching-based keyword-spotting to catch all the known diseases, we use our knowledge-base to ensure that we catch those diseases which are present in it.
Deep learning approach: Named entity recognition (NER) is a widely studied task in Natural Language Processing (NLP). Deep-learning-based transformer models like BERT8 have shown excellent performance on this task.
- Datasets: NCBI6 and BC5CDR7 are two well-known datasets that are commonly used as a benchmark for disease NER tasks, and we use a combination of them to train our models. However, NER datasets are not always available in other languages. It is also difficult to create good synthetic NER datasets. To overcome this, we use zero-shot learning.
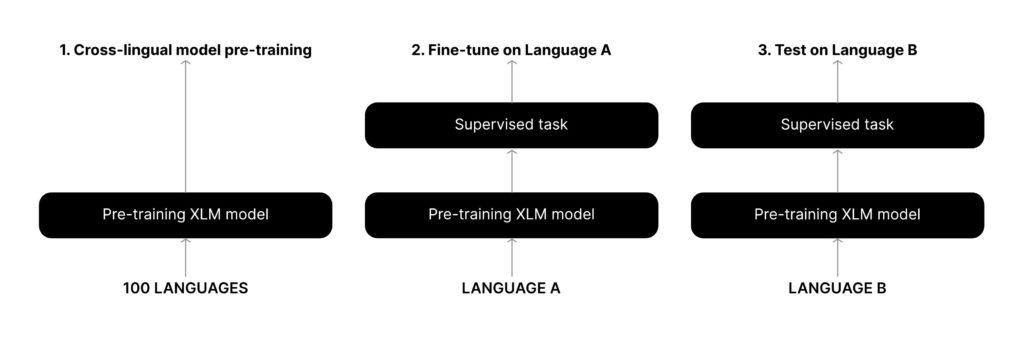
- Models: As mentioned earlier, for articles in English, we fine-tuned BioBERT on the datasets mentioned above for disease NER tasks. For other languages, we use the zero-shot capabilities of the XLM-R model. XLM-R is a transformer-based language model trained on a large corpus of multilingual datasets. Due to its unique pre-training steps, it is able to learn relations between different languages4. This allows the model to perform tasks in zero-shot fashion.
- Zero-shot learning: In zero-shot learning, a model is trained on one task and is directly used on some other (related) task without any fine-tuning. In our case, we fine tuned XLM-R on English disease NER tasks and used it to perform disease NER for other languages. We found this approach to be highly effective.
Location NER
For location NER, we take the traditional keyword-spotting approach, because location names are more or less constant. For this task, we created a knowledge-base of all the districts and sub-districts in India. New locations can be added to this knowledge-base as required.
For all the relevant articles that remain after the various filters we’ve used until this point in the process, we have gathered information such as disease and location tags. While this, in itself, will place the system on par with many existing systems used around the world, we decided to take this a step further and perform event extraction.
Event extraction
Mentions of multiple locations and multiple diseases can exist in the same article. How do we join them so that they form a meaningful event? In addition to this, we also have to extract other entities required to define an event, such as incidents, numbers, and incident types. We use a Question-Answering (QA) technique to accomplish this task. This is the most challenging—and most important—task within our system.
Each article is tagged with information related to diseases, locations, and dates. We use these tags as anchors to generate smart questions.
Smart questions
Since the quality of answers generated by QA models depends greatly on the quality of the questions asked, specific questions elicit specific outputs. Take, for instance, the example below:
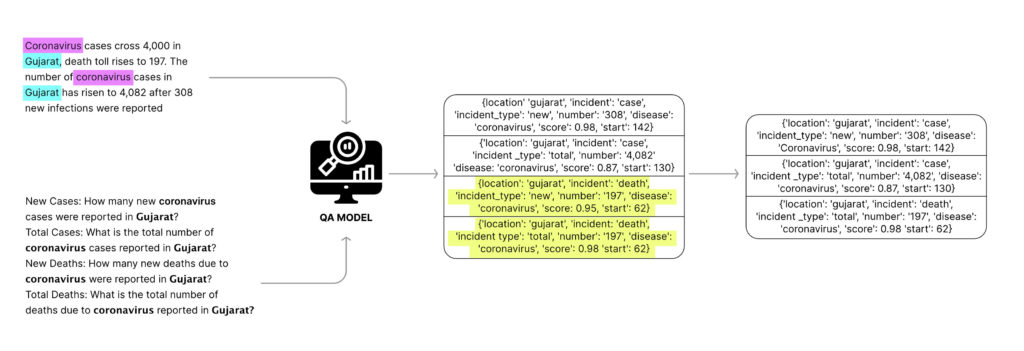
Article: Coronavirus cases cross 4,000 in Gujarat, death toll rises to 197, the total number of coronavirus cases in Gujarat has risen to 4,082 after 308 new infections were reported.
If we ask the question “how many new cases were reported?”, the QA model will struggle to find the appropriate answer because of the lack of a signal in the question itself. However, the question “how many new cases of coronavirus were reported in Gujarat?” was answered correctly by most QA models in our experiment as ‘4000’. Notice how using location and disease information in the question itself made the question very specific. We refer to such questions, anchored on disease and location, as ‘smart questions’.
Question-answering
The content in each news article serves as the context and we use a question-answering model to get answers to our smart questions. We apply a few filters to remove redundant answers, and then turn these answers into events. This results in the extraction of precise events from each article. We can understand this better by using the same article as an example.
Article: Coronavirus cases cross 4,000 in Gujarat, death toll rises to 197, the total number of coronavirus cases in Gujarat has risen to 4,082 after 308 new infections were reported.
In this article, we tag coronavirus as the disease and Gujarat as the location. Next, we generate smart questions anchored on these tags.
- How many new coronavirus cases were reported in Gujarat?
- What is the total number of coronavirus cases reported in Gujarat?
- How many new deaths were reported in Gujarat due to coronavirus?
- How many total deaths due to coronavirus were reported in Gujarat?
And so on.
To generate smart questions, we have created about 40–50 question templates, with slight variations. Each question template contains placeholders for locations and diseases, which are replaced with the location and disease found in each article. These questions can be divided into four categories:
- New cases
- Total cases
- New deaths
- Total deaths
Questions are created for each location-disease pair. The rationale behind this is that for each location-disease pair, there can only be one correct answer for each category. Once we have the answers for every question, we apply filters to arrive at the best answers. Along with the answer text, the model also outputs a score for that particular answer. The answers are then sent for filtering if, and only if, their score crosses a certain threshold. Using a threshold of 0.2 has given the best results on our test set.
Filtering
The following are a few of the filters that we use to discard redundant answers.
- Since there can only be one correct answer per category for every location-disease pair, we first consider answers with the maximum score from each category, for each unique pair.
- Next, we know that any substring of the article can be a part of one correct answer (at most). Thus, we retain the answer with the maximum score for every unique substring.
- Sometimes, numbers can be present in the form of words (“thirty”). We apply post-processing techniques to convert these words to numbers.
Once we filter all the answers, we create events from them. Answers to questions that belong to the ‘new cases’ category are assigned an incident as ‘case’, incident type as ‘new’, and so on. For example:
Question: How many new cases of coronavirus were reported in Gujarat?
Answer: 4000
Event: { location: Gujarat, disease: coronavirus, number: 4000, incident: case, incident_type: new }
Dataset
We use the SQuAD2.0 dataset, commonly used as a benchmarking dataset for QA tasks5, to train our QA models. We made a decision to use this dataset, even though is not specific to the task at-hand, as models trained on this dataset perform well on QA tasks in general. Another advantage of this dataset is that it contains “impossible to answer” questions. Such questions in the training dataset teach the model to refrain from answering when there is an insufficient number of signals in the context. This improves the overall precision of our system.
Since high quality QA datasets are not always available for other languages, the process is not as straightforward, and the zero-shot technique is used again.
Model
RoBERTa is a transformer-based model for English that is pre-trained on a large corpus of text, on language tasks such as Masked Language Modeling9. We fine-tuned the RoBERTa model on the SQuAD2.0 dataset, and used XLM-R again for other languages.
Evaluation
To evaluate our system, we created our own dataset consisting of news articles that were manually annotated with ground-truth events. Most of the articles present in our dataset had multiple ground-truth events, making the task even more challenging.
For evaluation, we use hard-matching, i.e., matches are considered as valid only if all the ground-truth entities and predicted events match. Using this strategy, we calculated the precision, recall, and F1-score of our system on this dataset. Along with English, we created this dataset for Hindi and Telugu, to evaluate the multilingual performance of our system.
Here are the results we observed for these languages.
| Language | Precision | Recall | F1 |
|---|---|---|---|
| English | 0.85 | 0.85 | 0.85 |
| Hindi | 0.84 | 0.79 | 0.82 |
| Telugu | 0.82 | 0.80 | 0.81 |
The system performs very well for all three languages, with a slightly lower performance for Hindi and Telugu. This is understandable, given the zero-shot nature of the models.
Deployment and impact
Once events have been finalised, they are inserted into a database, which then showcases them in our application.
We have integrated our AI solution within the Integrated Health Information Portal (IHIP), a portal managed by the Government of India, in which all the alerts from indicator- and event-based surveillance are logged. Raising an alert using our application directly logs it into the IHIP, and the alert is then distributed to the concerned authorities at the state and district levels. This ease of use has made it unnecessary to raise alerts manually, or communicate over SMS and email.
Our solution was deployed in April 2022 and has been used actively by the MSVC since then. Since its launch, it has crawled and analysed over 8 million articles and has raised hundreds of flags about potential events of significance. The MSVC has already raised over 200 alerts using our application.
Since our solution currently supports several Indian languages, it is able to capture local news reported in regional languages. We also provide translated versions of reported articles, so that reviewers are able to raise alerts based on articles written in a language other than those they may be familiar with (English, Hindi).
Our solution has received a lot of positive feedback from the officials using it for disease surveillance on a daily basis. It has greatly complemented their work and made the system more efficient. Now, less time is devoted to reading emails, which allows for more time to be dedicated towards analysing alerts and looking for meaningful trends. And though we are actively working on improving our solution, today, we are glad to say that our AI-powered solution is automating the event-based disease surveillance system in India.
References
- Linge, Jens & Verile, Marco & Tanev, Hristo & Zavarella, Vanni & Fuart, Flavio & van der Goot, Erik. (2011). Media Monitoring of Public Health Threats with MedISys.
- Freifeld, Clark C et al. “HealthMap: global infectious disease monitoring through automated classification and visualization of Internet media reports.” Journal of the American Medical Informatics Association : JAMIA vol. 15,2 (2008): 150-7. doi:10.1197/jamia.M2544
- Lee, Jinhyuk, et al. “BioBERT: A Pre-Trained Biomedical Language Representation Model for Biomedical Text Mining.” Bioinformatics, edited by Jonathan Wren, Sept. 2019. Crossref, https://doi.org/10.1093/bioinformatics/btz682.
- Conneau, Alexis et al. “Unsupervised Cross-lingual Representation Learning at Scale.” ACL (2020).
- Rajpurkar, Pranav et al. “Know What You Don’t Know: Unanswerable Questions for SQuAD.” ACL (2018).
- Doğan, Rezarta Islamaj et al. “NCBI disease corpus: a resource for disease name recognition and concept normalization.” Journal of biomedical informatics vol. 47 (2014): 1-10. doi:10.1016/j.jbi.2013.12.006
- Wei CH, Peng Y, Leaman R, Davis AP, Mattingly CJ, Li J, Wiegers TC, and Lu Z. Overview of the BioCreative V Chemical Disease Relation (CDR) Task. Proceedings of the Fifth BioCreative Challenge Evaluation Workshop, pp. 154-166
- [BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding](https://aclanthology.org/N19-1423) (Devlin et al., NAACL 2019)
- Liu, Yinhan et al. “RoBERTa: A Robustly Optimized BERT Pretraining Approach.” ArXiv abs/1907.11692 (2019): n. Pag.
- Choi, Hyunjin et al. “Analyzing Zero-shot Cross-lingual Transfer in Supervised NLP Tasks.” 2020 25th International Conference on Pattern Recognition (ICPR) (2021): 9608-9613.